RLP23 transfer enhances tomato resistance to multiple pathogens
The Arabidopsis receptor RLP23 recognizes the conserved immunogenic epitope nlp20, which is present in pathogenic fungi, oomycetes and certain bacteria12, making it a promising tool for crop improvement. To explore this potential, we generated a transgenic tomato line stably expressing RLP23 fused with GFP under the constitutive 35S promoter (p35S::RLP23–GFP). Western blot analyses confirmed successful expression of RLP23 (Supplementary Fig. 1a,b).
Compared with wild-type (WT) tomato plants, the transgenic line showed increased ethylene production in response to nlp20 stimulation (Fig. 1a). To assess whether RLP23-mediated recognition of nlp20 enhances immunity, we challenged the transgenic plants with the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000, the fungal pathogen Botrytis cinerea and the oomycete P. infestans. Notably, transgenic tomatoes displayed enhanced antibacterial immunity against Pst DC3000, despite the apparent absence of NLPs in this pathogen (Fig. 1b). We hypothesize that RLP23 may detect unknown bacterial ligands, similar to how RLP30 recognizes conserved cysteine-rich proteins from both fungi and oomycetes, as well as structurally unrelated bacterial ligands9. Additionally, RLP23-expressing plants showed significantly smaller disease lesions upon infection with both B. cinerea and P. infestans compared with WT plants (Fig. 1c,d), confirming that RLP23 expression confers broad-spectrum resistance to diverse pathogens in tomato.
a, Ethylene accumulation in WT and transgenic tomato plants expressing RLP23, RLP23 with AtSOBIR1, RLP23/CTCf-9 or RLP23/CTEIX2, measured after 4 h of treatment with 2 µM nlp20. b, Bacterial growth of Pst DC3000 in WT and transgenic tomato plants, quantified as CFU in leaf extracts 3 days after inoculation. c,d, Lesion diameters on WT and transgenic tomato leaves infected with B. cinerea (c) or P. infestans (d), assessed 2 days after drop inoculation. e,f, Total weight (e) and number (f) of mature fruits collected per tomato plant. Data in a, e and f are presented as the mean ± s.d. (n = 6 for a; n = 8 for e and f). For b–d, data are displayed as box plots (center line, median; bounds of box, the first and third quartiles; whiskers, 1.5 times the interquartile range; error bar, minima and maxima; n = 20 for b; n = 18 for c and d). Data were analyzed using a two-way analysis of variance (ANOVA) with Tukey’s test. Statistically significant differences from WT are indicated (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001). Each experiment was repeated three times with consistent results.
To further explore whether coexpression of RLP23 with the adaptor protein AtSOBIR1 could boost immunity, we generated a transgenic tomato line expressing both RLP23 and AtSOBIR1. Coexpression resulted in a modest increase in nlp20-induced ethylene production compared with RLP23 expression alone (Fig. 1a). However, AtSOBIR1 expression also caused dwarfism, likely because of autoimmune activation (Supplementary Fig. 1a). This phenotype contrasts with N. tabacum, where coexpression of RLP30 and AtSOBIR1 did not affect plant growth9, suggesting species-specific differences in AtSOBIR1-mediated immune regulation. While coexpression with AtSOBIR1 slightly enhanced immune signaling, its negative impact on growth limits its practical application for crop improvement. Overall, these results demonstrate that stable ectopic expression of RLP23 in tomato promotes broad-spectrum resistance to destructive bacterial, fungal and oomycete pathogens.
Full RLP functionality in heterologous plants depends on the CT domains
RLPs contain a short C-terminal intracellular domain (IC domain) (Supplementary Fig. 2) but its role in immune signaling remains unclear. To investigate its contribution to RLP-mediated immunity, we introduced a truncated RLP23 variant lacking the IC domain (RLP23ΔIC) into the Arabidopsis rlp23-1 mutant. However, deletion of the IC domain did not impair ethylene accumulation in response to nlp20 treatment (Supplementary Fig. 3a,b). Surprisingly, transient expression of RLP23ΔIC in N. benthamiana resulted in a significantly weaker ethylene response to nlp20 compared with full-length RLP23 (Supplementary Fig. 3c,g), suggesting that the IC domain has a species-dependent role in immune activation.
We also examined RLP30, an SCP receptor with a short IC domain of 23 aa9. In an N. benthamiana line lacking the SCP receptor RE02 (ΔRE02), expression of full-length RLP30 reconstituted SCP-triggered ethylene production, whereas the truncated variant lacking the intracellular structure (RLP30ΔIC) failed to restore the responses to RLP30 WT levels (Supplementary Fig. 3d,h).
Interestingly, the IC domains of RLPs harbor putative phosphorylation sites—four in RLP30 (T761, T783, T784 and S785) and two in RLP23 (S872 and Y873) (Supplementary Fig. 2b). To assess their functional relevance, we generated phospho-dead mutants by substituting these residues with alanine or phenylalanine (T761A;T783A;T784A;S785A in RLP304mut and S872A;Y873F in RLP232mut). Immune responses triggered by both SCP and nlp20 remained unchanged in N. benthamiana plants expressing the mutant receptors (Supplementary Fig. 3e,f,i,j), indicating that RLP-mediated immunity operates independently of IC domain phosphorylation.
In addition to the IC domain, the CT region of RLPs contains transmembrane (TM) and juxtamembrane (JM) domain (Supplementary Fig. 2). To assess their contribution to immune signaling, we generated two RLP23 variants: one lacking both TM and IC and another with a complete CT deletion (CT, IC + TM + JM) (Supplementary Fig. 4a). Ethylene accumulation induced by nlp20 was comparable in N. benthamiana expressing RLP23ΔIC + TM and RLP23ΔIC, whereas additional deletion of the JM domain further compromised immune responses (Supplementary Fig. 4b). Similar results were obtained with RLP30 CT truncations (Supplementary Fig. 4c). These findings highlight the essential role of the CT domains in RLP-mediated immunity in heterologous plant systems.
CT swaps modulate RLP functionality
The reduced activity upon CT deletions of RLP23 and RLP30 in N. benthamiana underscores the critical role of the C terminus in heterologous expression systems. To assess whether CT domains from solanaceous RLPs could enhance RLP23 functionality in the Solanaceae family, we selected two well-characterized tomato RLPs, EIX2 and Cf-9, as CT donors because of their overall structural similarity to RLP23 (Supplementary Fig. 2). EIX2 detects ethylene-inducing xylanase, while Cf-9 recognizes the Cladosporium fulvum effector protein Avr9 (refs. 13,14).
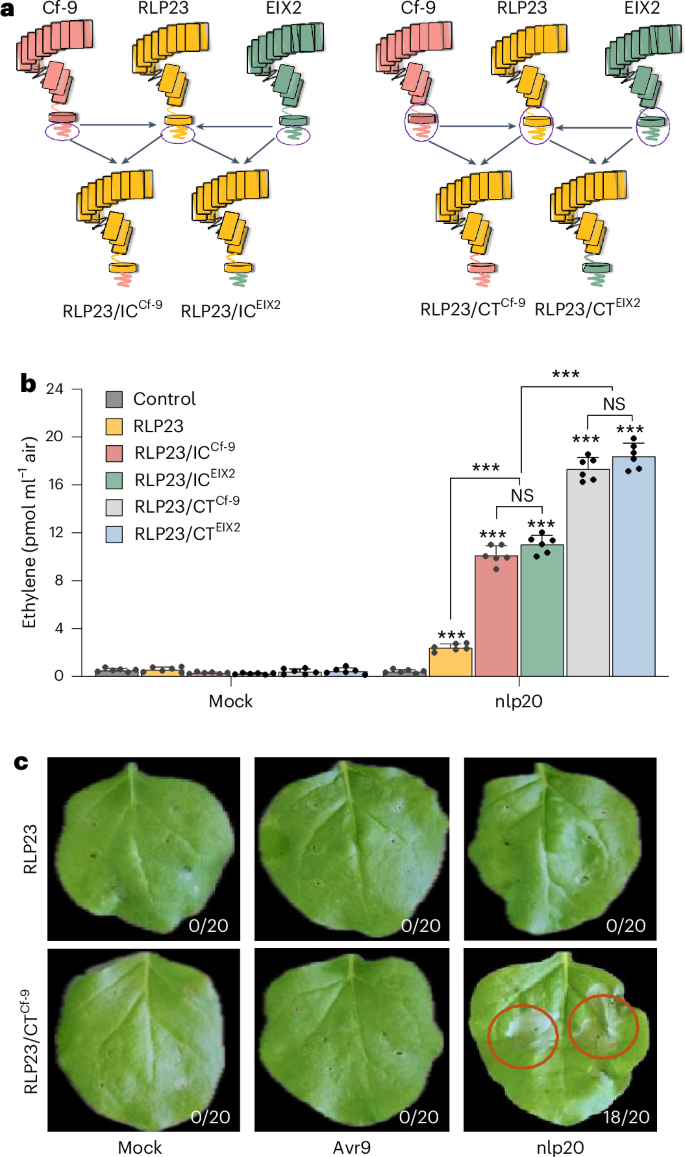
We constructed chimeric receptors by swapping the IC or CT domains of RLP23, EIX2 and Cf-9 (Fig. 2a and Supplementary Figs. 5 and 6). Expression of RLP23/ICEIX2 and RLP23/ICCf-9 in N. benthamiana resulted in a fourfold increase in ethylene production in response to nlp20 treatment compared with WT RLP23 (Fig. 2b), indicating that the IC domains from solanaceous RLPs enhance RLP23-mediated immune responses.
a, Schematic representation of RLP23, Cf-9 and EIX2, along with their chimeric variants featuring reciprocal IC or CT domain swaps. b, Ethylene accumulation in N. benthamiana leaves transiently expressing GFP (control), RLP23–GFP, RLP23/ICCf-9–GFP, RLP23/ICEIX2–GFP, RLP23/CTCf-9–GFP or RLP23/CTEIX2–GFP, measured 4 h after treatment with water (mock) or 2 µM nlp20. Data are presented as the mean ± s.d. (n = 6) and were analyzed using a two-way ANOVA with Tukey’s test. Statistically significant differences from control plants are indicated (***P ≤ 0.001; NS, not significant). c, Cell death development in N. benthamiana leaves transiently expressing RLP23–GFP or RLP23/CTCf-9–GFP, 72 h after infiltration with water (mock), 20 µM nlp20 or 100 µl of apoplastic fluid with Avr9. The number of necrotic leaves is presented relative to the total number of infiltrated leaves. Each experiment was repeated three times with similar results.
Substitution of the entire RLP23 CT region with that of tomato RLPs further amplified its immune function in N. benthamiana, measurable by elevated ethylene production in response to nlp20 (Fig. 2b). A similar enhancement was observed upon editing the full CT region of RLP30, which led to stronger SCP-induced immune responses in ΔRE02 plants expressing RLP30/CTEIX2 or RLP30/CTCf-9 (Supplementary Fig. 5b). In contrast, replacing the CT domains in EIX2 and Cf-9 with that of RLP23 significantly reduced their respective responses to EIX and Avr9 (Supplementary Fig. 6b,c), demonstrating that the CT domains contribute to receptor-specific immune signaling.
Interestingly, RLP23/CTCf-9 expression induced nlp20-triggered cell death, a hallmark of ETI that was not previously observed in any plant following nlp20 treatment (Fig. 2c). Conversely, Cf-9/CTRLP23 did not produce the cell death response usually triggered by Avr9 (Supplementary Fig. 6e), indicating that the C terminus has a key role in regulating immune signaling outcomes. These findings highlight the functional importance of the C terminus in modulating RLP activity, enhancing immune responses and influencing cell death phenotypes in heterologous expression systems.
Disease resistance in tomato is enhanced by RLP23 CT editing without yield loss
Building on the enhanced RLP23 functionality observed in N. benthamiana, we explored whether RLP23 chimeric receptors could improve disease resistance in tomato. The RLP23/CTEIX2 and RLP23/CTCf-9 constructs were stably introduced into the tomato cultivar Moneymaker, which does not naturally respond to nlp20. Consistent with the results from N. benthamiana, transgenic tomato lines expressing the RLP23 hybrid receptors showed a substantial increase in nlp20-induced ethylene production and enhanced disease resistance (Fig. 1a–d). Compared with WT and RLP23-expressing plants, RLP23/CTEIX2 and RLP23/CTCf-9 transgenic tomatoes exhibited significantly reduced bacterial growth after Pst DC3000 infection (Fig. 1b) and developed smaller disease lesions when challenged with the necrotrophic fungus B. cinerea (Fig. 1c) or the oomycete P. infestans (Fig. 1d).
The tradeoff between defense and growth has long been a key challenge in crop breeding15. In contrast to the dwarfism observed in plants coexpressing RLP23 with AtSOBIR1, tomato lines carrying RLP23/CTEIX2 and RLP23/CTCf-9 showed no developmental defects, with growth patterns comparable to those of WT and RLP23-expressing plants (Supplementary Fig. 1a). Importantly, the enhanced resistance conferred by the engineered RLP23 receptors did not affect fruit yield. Both transgenic and WT tomato plants produced similar numbers and total weight of mature fruits (Fig. 1e,f). These findings demonstrate that RLP engineering can significantly enhance broad-spectrum resistance in tomato without compromising yield.
Interaction of RLP23 with SOBIR1 is modulated by the CT domain
To understand how tomato RLP CT domains enhance RLP23 functionality in Solanaceae, we examined their impact on immune signaling. Consistent with the elevated nlp20-triggered ethylene levels observed in RLP23-expressing plants, we also detected significant upregulation of immune-related gene transcription upon nlp20 treatment. Genes such as PR1a and RIPK were strongly induced, with even higher expression levels in transgenic tomato lines expressing the chimeric RLP23/CTEIX2 (Supplementary Fig. 7a). Notably, these lines accumulated transcripts of genes typically associated with ETI, such as EDS1, NRC2 and NRC4a, in response to nlp20 but not to treatment with flg22 (Supplementary Fig. 7a,b). These findings suggest that the immune response triggered by RLP23 variants in Solanaceae involves elements known to function in ETI pathways.
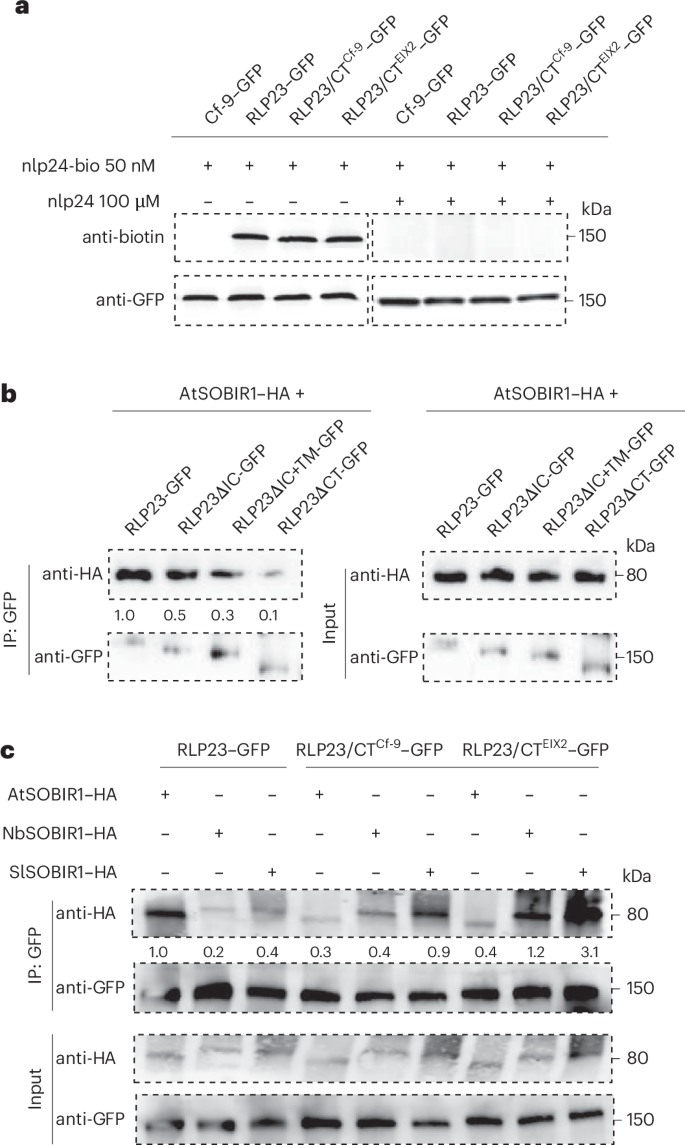
To assess whether ligand-binding capacities were altered in the RLP23 chimeric proteins, we tested their interaction with biotinylated nlp24 (nlp24-bio), a functional analog of nlp20 that includes a C-terminal 4-aa extension. All tested variants, including WT RLP23, RLP23/CTEIX2 and RLP23/CTCf-9, associated similarly with nlp24-bio. Excess of untagged nlp24 effectively displaced nlp24-bio from all RLP23 variants (Fig. 3a), indicating that CT domain replacement does not affect the ligand-binding properties of RLP23. Thus, the enhanced immune response is unlikely to be because of altered ligand recognition.
a, Ligand-binding assay in N. benthamiana leaves transiently expressing indicated GFP-tagged RLP variants. Leaves were treated with biotinylated nlp24 (nlp24-bio) in the presence (+) or absence (−) of unlabeled nlp24, followed by coimmunoprecipitation using a GFP-Trap. b, Coimmunoprecipitation assay in N. benthamiana leaves coexpressing GFP-tagged RLP23 variants with AtSOBIR1–HA. Leaf protein extracts (input) were subjected to coimmunoprecipitation with GFP beads (IP:GFP) and immunoblotting using indicated antibodies. c, Coimmunoprecipitation assay in N. benthamiana leaves coexpressing GFP-tagged RLP23 variants with AtSOBIR1–HA, NbSOBIR1–HA, or SlSOBIR1–HA. Leaf protein extracts (input) were subjected to coimmunoprecipitation with GFP beads (IP:GFP) and immunoblotting using tag-specific antibodies. Protein levels were quantified relative to AtSOBIR1–HA precipitated by RLP23–GFP, set as 1 (b,c). Representative results from three independent experiments are shown.
In addition, we evaluated the roles of RLP23 CT region in mediating interaction specificity with the adaptor kinase SOBIR1. Coimmunoprecipitation assays revealed that deletion of the IC or IC + TM domains markedly reduced RLP23–SOBIR1 interaction, while removal of the entire C terminus nearly abolished binding (Fig. 3b). Interestingly, chimeric receptors RLP23/CTEIX2 and RLP23/CTCf-9 showed stronger association with SOBIR1 from N. benthamiana and tomato than with AtSOBIR1 (Fig. 3c and Supplementary Fig. 8). While WT RLP23 preferentially associated with AtSOBIR1, the chimeric variants exhibited the strongest association with SlSOBIR1 (Fig. 3c and Supplementary Fig. 8). These findings highlight the critical role of the C terminus in modulating RLP–SOBIR1 interactions and demonstrate that its targeted engineering can improve RLP23’s adaptation and integration into the immune signaling networks of heterologous plants.
RLP23 expression in rice confers recognition of NLPs from blast fungi
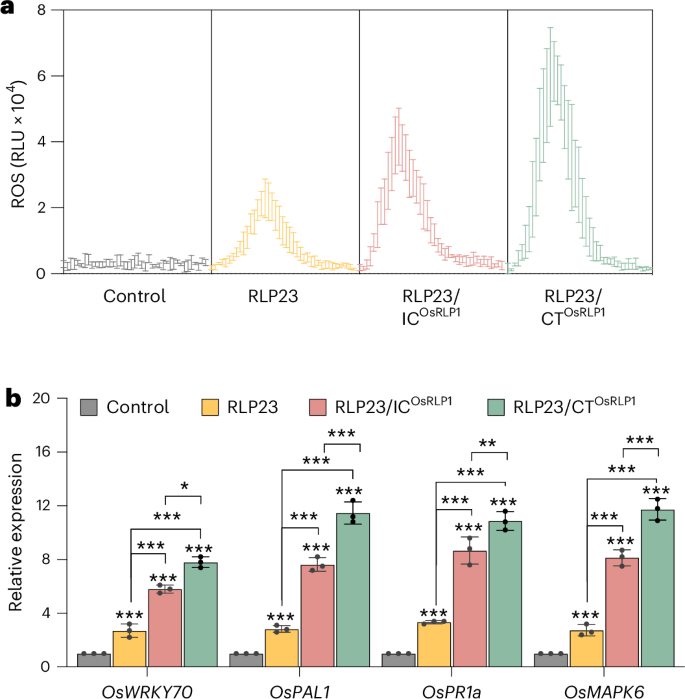
Given that Arabidopsis RLP23 conferred resistance in dicot plants such as tomato and tobacco (Figs. 1 and 2), we next investigated its potential to improve immunity in monocot crops. The rice blast fungus Magnaporthe oryzae encodes the NLP protein MoNLP1, which contains the conserved immune epitope nlp20 (ref. 16). However, rice does not naturally recognize this protein (Fig. 4a). To determine whether RLP23 can mediate MoNLP1 recognition in rice, we transiently expressed the p35S::RLP23–GFP construct in rice protoplasts. Upon MoNLP1 treatment, RLP23 expression triggered a significant accumulation of reactive oxygen species (ROS) (Fig. 4a). Moreover, MoNLP1-treated RLP23-expressing rice protoplasts showed significant upregulation of defense-related genes, including OsWRKY70, OsPAL1, OsPR1a and OsMAPK6 (Fig. 4b). These results indicate that RLP23 enables MoNLP1 recognition and activates defense responses in rice.
a, ROS production in rice protoplasts expressing GFP (control), RLP23–GFP, RLP23/ICOsRLP1–GFP or RLP23/CTOsRLP1–GFP, following treatment with 5 µM purified MoNLP1. Data are presented as relative light units (RLU) over 2 h. b, RT–qPCR analysis of rice protoplasts expressing GFP (control) or GFP-tagged RLP23 variants, treated with 2 µM MoNLP1 for 24 h. Expression levels of the indicated marker genes were quantified using gene-specific primers, normalized to OsActin transcript levels and presented relative to the control (set to 1). Data are presented as the mean ± s.d. (n = 3) and were analyzed using a two-way ANOVA with Tukey’s test. Statistically significant differences from the control are indicated (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001). Each experiment was performed three times with similar results.
To optimize RLP23 functionality in rice, we designed hybrid receptors RLP23/ICOsRLP1 and RLP23/CTOsRLP1 by fusing RLP23 with corresponding domains of rice OsRLP1, a key component of antiviral immunity in rice17. Compared with WT RLP23, expression of RLP23/ICOsRLP1 in rice protoplasts led to further increased ROS production and stronger defense gene induction upon MoNLP1 treatment, while further extending the modification to encompass the full C terminus amplified these immune responses (Fig. 4). However, fusion with the C terminus of tomato RLP EIX2 failed to boost RLP23 activity in rice, emphasizing the species-specific nature of CT domain compatibility for cross-species receptor function (Supplementary Fig. 9a). These findings underscore the potential of RLP engineering to improve disease resistance in monocot crops such as rice.
Heterologous expression of Arabidopsis RLP23 enhances fungal resistance in poplar
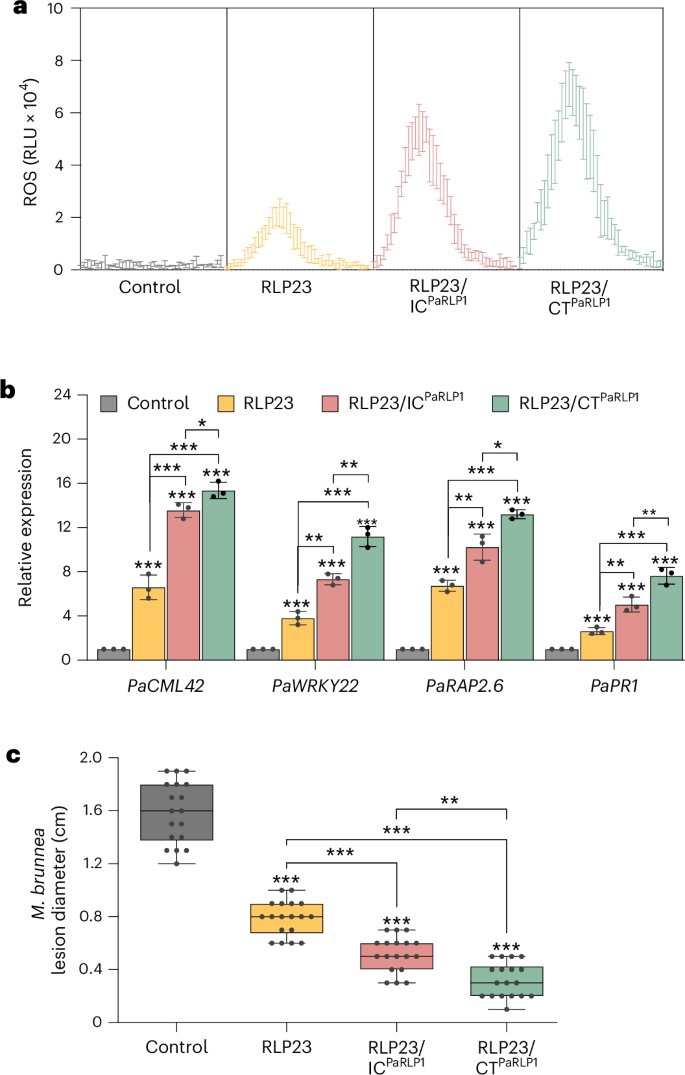
RLP23 is specific to the Brassicaceae family, while NLP proteins are widely conserved among fungal pathogens, including Marssonina brunnea, the causal agent of black spot disease in the economically important tree species poplar18. To explore the potential of cross-species RLP transfer for tree improvement, we used the NLP-type protein MbNLP1 from M. brunnea, which is recognized by RLP23 but not by poplar18 (Fig. 5a). MbNLP1 contains a conserved 24-aa immunogenic fragment (nlp24MbNLP1), which induces ROS accumulation and upregulates defense gene expression in poplar leaves transiently expressing RLP23 (Fig. 5a,b). Importantly, ectopic expression of RLP23 conferred fungal resistance, as evidenced by reduced disease symptoms in poplar leaves compared with GFP-expressing controls (Fig. 5c).
a, ROS accumulation in poplar leaves 48 h after transient expression of GFP (control) or the indicated constructs, followed by treatment with 2 µM nlp24MbNLP1. ROS levels were recorded over 2 h and are presented as RLU. b, RT–qPCR analysis of poplar leaves expressing GFP (control), RLP23, RLP23/ICPaRLP1 or RLP23/CTPaRLP1 for 48 h, followed by treatment with 1 µM nlp24MbNLP1 for 24 h. Expression levels of the indicated genes were determined using gene-specific primers, normalized to PaEF1α transcript levels and presented relative to the control (set to 1). Data are presented as the mean ± s.d. (n = 3). c, Lesion diameter measured 3 days after inoculation with M. brunnea in poplar leaves transiently expressing GFP (control) or the indicated RLP–GFP fusion constructs. Data (n = 18) are shown as box plots (center line, median; bounds of box, the first and third quartiles; whiskers, 1.5 times the interquartile range; error bar, minima and maxima). All data were analyzed using a two-way ANOVA with Turkey’s test. Statistically significant differences from control plants are indicated (*P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001). Shown is one representative experiment of three with similar results.
To further enhance RLP23 function in poplar, we engineered the chimeric receptors RLP23/ICPaRLP1 and RLP23/CTPaRLP1 by fusing the IC domain or the entire C terminus of poplar RLP1 with RLP23 (ref. 19). Similar to tomato and rice, poplar leaves expressing RLP23/CTPaRLP1 showed higher ROS accumulation and more pronounced defense gene expression in response to nlp24MbNLP1 compared with expression of WT RLP23 or the IC domain-swapped variant (Fig. 5a,b). These elevated immune responses translated into significantly enhanced antifungal resistance (Fig. 5c), demonstrating that RLP engineering can be effectively applied to tree breeding, offering a promising strategy for improving disease resistance in perennial crops.